A new genetic lineage of Asparagopsis taxiformis (Rhodophyta) in the Mediterranean Sea: As the DNA barcoding indicates a recent Lessepsian introduction
- 1Department of Marine Biology, the Leon H. Charney School of Marine Sciences, University of Haifa, Haifa, Israel
- 2Department of Marine Biology, Israel Oceanographic & Limnological Research, Ltd. (PBC), Haifa, Israel
Asparagopsis taxiformis (Delile) Trevisan is a red marine macroalga (Bonnemaisoniales, Rhodophyta) with high invasive potential and broad worldwide distribution. In the Mediterranean Sea, A. taxiformis was reported before the opening of the Suez Canal and is comprised of two different cryptic lineages, named L2 and L3. As for the Israeli Mediterranean Sea (IMS), A. taxiformis benthic populations have seemingly expanded with several large seasonal blooms recorded in recent years. However, neither ecology nor molecular substantial studies have been conducted for this particular geographical area. Increasing sampling intensity and geographical coverage may reveal new lineages or indicate human-mediated spread routes not only for A. taxiformis but for macroalgae in general. This approach is particularly important in areas such as the eastern Mediterranean Sea, which experiences intense biological invasion on a global scale. In this study, randomly samples specimens (n = 30) of A. taxiformis and preserved herbarium samples (n = 4) collected from the IMS in the past, were all barcoded and taxonomically identified using three molecular genetic markers (LSU, cox2-3 spacer, and rbcL). We found a cryptic lineage 4 (L4) of A. taxiformis first reported here for the Mediterranean Sea, and previously described for the western Indo-Pacific and Hawaii. Herbarium samples confirmed the presence of L4 as early as 2013. Comparative assessment of cox2-3 spacer marker indicates 100% similarity to sequenced L4 samples from Egypt in the Red Sea. The IMS cox2-3 spacer sequences differed from previously sequenced samples from the Mediterranean Sea by 2.3% and 3.9% bp, compared to L3 and L2 Mediterranean populations, respectively. Morphological inspections indicate monoecious L4 gametophytes which are larger than the L4 population reported previously from Hawaii. Altogether, our results strongly indicate a Lessepsian migration route for A. taxiformis L4 with yet unknown consequences for the local marine ecosystems.
Introduction
Marine coastal habitats in the Mediterranean Sea are severely biologically invaded ecosystems (Boudouresque and Verlaque, 2002). This is largely due to human-mediated introduction of non-native species through various vectors such as ballast waters of ships, regional aquaculture activities, and by natural dispersal via the Suez Canal (Lessepsian invasion) (FD, 1978; Grosholz, 2002; Rilov and Crooks, 2009; Katsanevakis et al., 2014). The total number of non-indigenous seaweed species (NISS) reported for the whole Mediterranean Sea is somewhat variable, which is not surprising given the complex cryptic nature of some of the species involved. The date of introduction and total numbers are subject to conjecture; nonetheless, agreed estimates may vary from as high as 148 species (as calculated for the year 2022 using the suggested 2–3 new immigrants per year by Zenetos et al. (2012), down to 118 species (Verlaque et al., 2015). The arrival of NISS is more intense in the eastern Mediterranean Sea since there is a consensus that the major vector of the introduction of macroalgae is of Lessepsian origin (Boudouresque and Verlaque, 2005; Zenetos, 2010; Zenetos et al., 2012; Verlaque et al., 2015; Romero et al., 2016).
In the last decades, the Israeli Mediterranean coast has become the host of several NISS, primarily in subtidal areas, and witnessed intense onshore drifts (Israel et al., 2019). Examples are the green alga Codium parvulum (Bory ex Audouin) P.C.Silva (Israel et al., 2010), the red alga Galaxaura rugosa (J.Ellis & Solander) J.V.Lamouroux (Hoffman et al., 2008), the brown algae Stypopodium schimperi (Kützing) Verlaque & Boudouresque (Verlaque and Boudouresque, 1991; Einav and Israel, 2009), Dictyota sp. (unpublished observations) and Lobophora lessepsiana C.W.Vieira (Vieira et al., 2019), and many others which are unaccounted for. In this context, the red alga Asparagopsis taxiformis (Delile) Trevisan, allegedly introduced into the Mediterranean Sea in 1831 (Verlaque et al., 2015) was hardly noticeable in the Israeli shores until about a decade ago. A. taxiformis is highly invasive and has therefore been underpinned as one of the ‘worst invasive alien species threatening marine biodiversity in Europe’ (EEA, 2007), and further listed within the 100 ‘worst invasive seaweeds in the Mediterranean Sea’ (Streftaris and Zenetos, 2006)
A. taxiformis is regarded as a cryptic complex with high diversity and cosmopolitan distribution from warm-temperate to tropical marine environments (Harvey, 1849; Price et al., 1986; Boni and Hawkes, 1987; Silva et al., 1996; Zanolla et al., 2022). The heteromorphic life cycle of A. taxiformis includes the erect gametophyte and a filamentous tetrasporophyte (Mairh, 1977; Guiry and Dawes, 1992). The gametophytic stage is highly branched, colored pink to red reaching up to 40 cm tall and commonly occurs on rocky substrates, or as epiphytes (Rojas et al., 1982; Boni and Hawkes, 1987). The tetrasporophytic stage, mistakenly referred in the past as a different species (Falkenbergia hillebrandii (Bornet) Felkenberg) (Chihara, 1961), consists of microscopic three-cell row filaments arranged in a pompon morphology up to 2 cm in diameter. Tertrasporophytes can be found free-floating or attached to other algae and are capable of dispersion by flotation. The high vegetative reproduction potential of the tetrasphorophyte stage serves as a prolific propagator for expanding population and invasion to new habitats (Chihara, 1961; Mairh, 1977; Zanolla et al., 2022).
To date, worldwide, six mitochondrial lineages have been genetically distinguished in the A. taxiformis populations (Andreakis et al., 2007; Sherwood, 2008; Dijoux et al., 2014; Andreakis et al., 2016; Zanolla et al., 2022). The cox2-3 spacer is considered the ideal marker for this linage separation differing in 6-23 bp out of the 338 bp of cox2-3 spacer marker (Dijoux et al., 2014; Andreakis et al., 2016; Kurihara et al., 2016). Based on this molecular tool, lineage 1 (L1) was described for the Pacific region, whereas a L2 additionally described for the Indo-Pacific (Andreakis et al., 2007; Dijoux et al., 2014), the Mediterranean Sea (Andreakis et al., 2007), and North Atlantic regions (Dijoux et al., 2014). L3 is known for the Mediterranean Sea (Andreakis et al., 2007) and also described for the western Atlantic (Andreakis et al., 2007), the Canary Islands and South Africa (Bolton et al., 2011). L4 can be found in Hawaii and the western Indo-Pacific (Sherwood, 2008) while L5 is found in Western Australia and the southern Indo-Pacific Ocean (Dijoux et al., 2014; Andreakis et al., 2016). Recently, L6, considered endemic to Australia was described by Andreakis et al. (2016). As mentioned before, A. taxiformis was first documented in the Mediterranean Sea before the opening of the Suez Canal, and its current distribution is likely the result of several introduction events including possible Lessepsian migration (Andreakis et al., 2004; Ní Chualáin et al., 2004; Andreakis et al., 2007; Dijoux et al., 2014). To date, only two cryptic lineages, L2 and L3, have been described for the Mediterranean Sea (Zanolla et al., 2022). Within the Israeli Mediterranean Sea (IMS), A. taxiformis has become abundant at 0-15 m deep, generally attached to hard bottoms. The presence of A. taxiformis along the Israeli coasts has been documented only twice (Lipkin, 1962; Einav and Israel, 2008), and no ecological or molecular biodiversity were reported for the local populations in the past. Field collections of specimens preserved at the Seaweed Herbarium of the Israel Oceanographic & Limnological Research, Ltd. (IOLR) (www.seaweedherbarium.com) confirm the expansion of this species during the last decade.
Global human activities are responsible for the spreading of marine organisms and the changes imposed on marine ecosystems on a worldwide scale. To detect and follow those changes, surveys on a local scale are necessary, having significant implications both for marine conservation and coastal ecosystem management (Bickford et al., 2007; Andreakis and Schaffelke, 2012). In this study we investigate for the first time the A. taxiformis populations of the IMS, integrating a DNA barcoding approach with morphological tools. In order to gain a better understanding of the distribution and origin of the IMS A. taxiformis, three genetic markers, nuclear LSU, mitochondrial cox2-3 spacer and plastid rbcL were used.
Materials and methods
Seaweed collection
Gametophytes (n= 28) and tetrasphorophyte (n= 2) of A. taxiformis were collected from five sites along the northern coast of the Israeli Mediterranean Sea (IMS) (Bat Galim 32°50’11.1”N 34°58’40.6”E, Tel Shikmona 32°49’33.8”N 34°57’16.8”E, Achziv 33°03’21.9”N 35°06’06.9”E, Rosh Hanikra 33°05’18.0”N 35°06’20.1”E and Sdot Yam 32°29’29.0”N 34°53’02.6”E) (Figure 1) between July 2020 and December 2021. At each site, thalli pieces from three different specimens situated at least three m apart from each other, were placed in 1 ml buffer lysis solution (40 mM EDTA, 50 mM Tris pH 8.3, and 0.75 M sucrose), transported to the laboratory and kept at -80°C until DNA extraction. Occasionally, fewer replicates were collected from sites with low algal density (Table S1). Part of the collected material was kept in a cooler for binocular observations and measurements of morphological parameters, that included: thallus length and maximum width, basal diameter of the main axis, height where branching begins and the length of the branchlets and their basal diameter. Four herbarium samples (www.seaweedherbarium.com) collected between 2013 - 2018 were also analyzed.
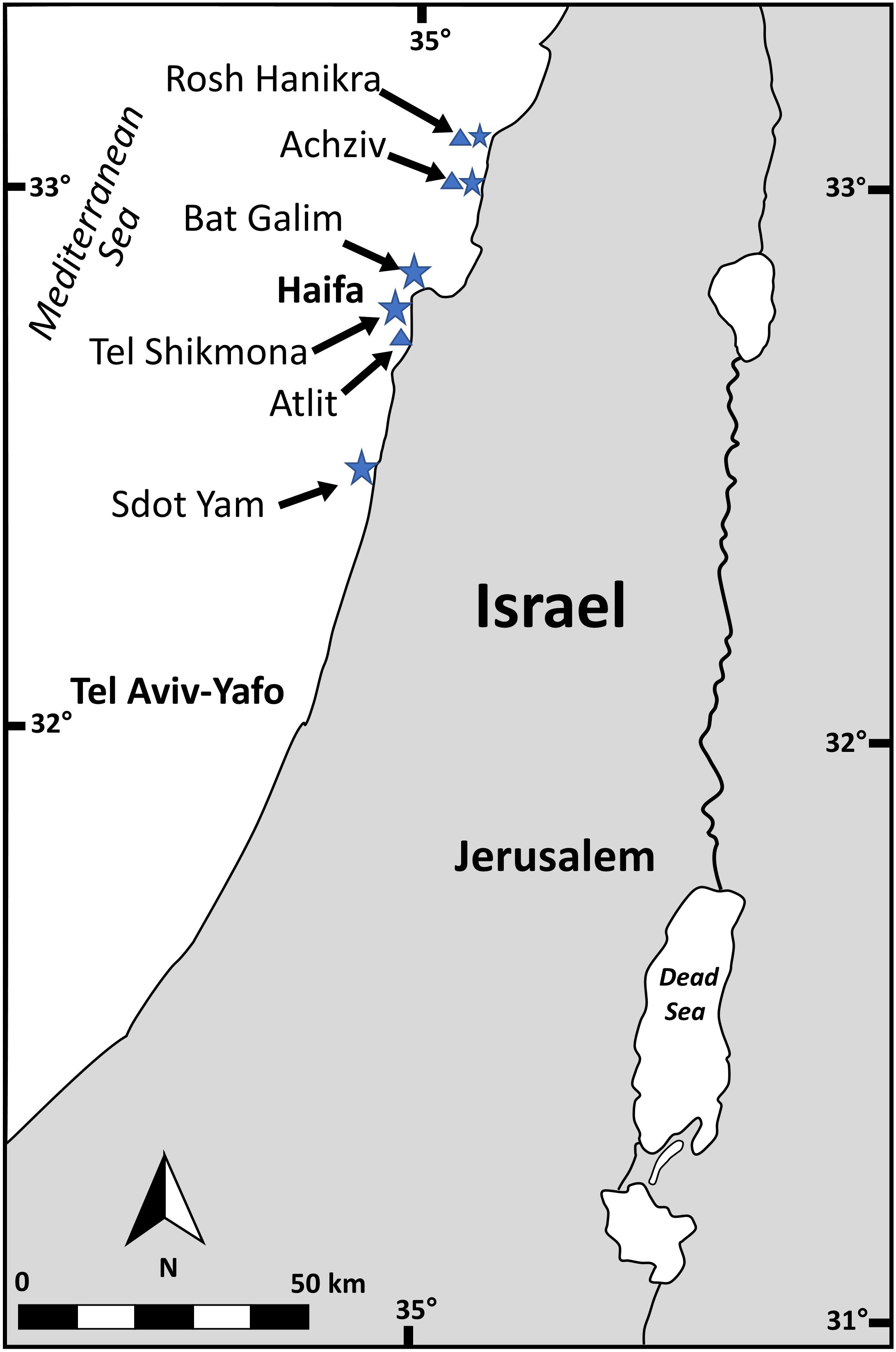
Figure 1 Sampling site across the Israeli Mediterranean Sea (IMS). Stars indicate fresh specimens sampling sites and triangles indicate herbarium sampling sites.
DNA extraction and molecular analyses
About 100 mg of fresh macroalgae biomass were placed in 1 ml buffer lysis solution (ISOLATE II Plant DNA Kit, Bioline) and homogenized using a small plastic pestle. DNA extraction from herbarium samples processed similarly as the fresh samples with an additional bead beater step to brake the cells walls. Genomic DNA was extracted as described in the ISOLATE II Plant DNA Kit. Quantity and quality of DNA were examined using a nanodrop (NANODROP 2000c Spectrophotometer, Thermo Scientific, USA).
Molecular lineages identification
Three barcoding markers, nuclear LSU, mitochondrial cox2-3 spacer and plastid rbcL, were amplified by polymerase chain reaction (PCR) for lineage identification (Zuccarello and Succursale, 1999; Andreakis et al., 2004; Dijoux et al., 2014). Primer information, such as locus names, nucleotide sequences, and references are provided in Table 1. The following PCR conditions were used for both LSU and rbcL PCR reaction: initial denaturation at 94°C for 5 min, followed by 30 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 90 s, with a final elongation step of 72°C for 10 min. For the mitochondrial cox2–3 spacer the following PCR conditions were used: initial denaturation at 94°C for 4 min, followed by 5 cycles of 93°C 45°C, and 72°C for 60 s each, followed by 30 cycles of 93°C, 50°C, and 72°C for 30 s each, with a final elongation step of 72°C for 10 min as described by Zuccarello and Succursale (1999). All PCR reactions were run in 50 µL containing 1 µL each of forward and reverse primers (10µM), 25 µL of ready Mix (Bioline Meridian Life Science Inc.), 1 µL template DNA, 20 µL of PCR Grade H2O and 2 µL of bovine serum albumin (BSA). PCR fragments were sequencedby Macrogen Europe(Macrogen;Europe BV, Amsterdam, Netherlands).
Sequence analysis
DNA sequences from this study and sequences deposited from GenBank were aligned using BioEdit v4.8.5 (Hall, 1999). The evolutionary history tree was inferred using the UPGMA method (P.H.A and Sokal, 1973) in MEGA- X software (Kumar et al., 2018). The evolutionary distances were computed using the p-distance method (No Nei and Kumar, 2000).
Results
Overall, 30 specimens of Asparagopsis taxiformis, 28 gametophytes and 2 tetrasphorophyte were collected in five sites along the northern Israeli coastline, from July 2020 until December 2021. Sampling efforts in southern shores, which are largely composed of sandy bottoms, indicated the absence of this species in soft bottoms. Morphological features of mature samples collected from the dense Rosh Hanikra site populations are shown in Table 2. IMS samples showed cystocarps (female reproductive structures) and spermatangia (male reproductive structures) in the majority of the samples examined (Figures 2, 3). Specifically, out of the 28 collected gametophytes, 20 were found to be monoecious, and no reproductive organs were observed for the rest.

Table 2 Morphological characters measured in gametophytes of A. taxiformis collected from the Rosh Hanikra site in Nov 2021.
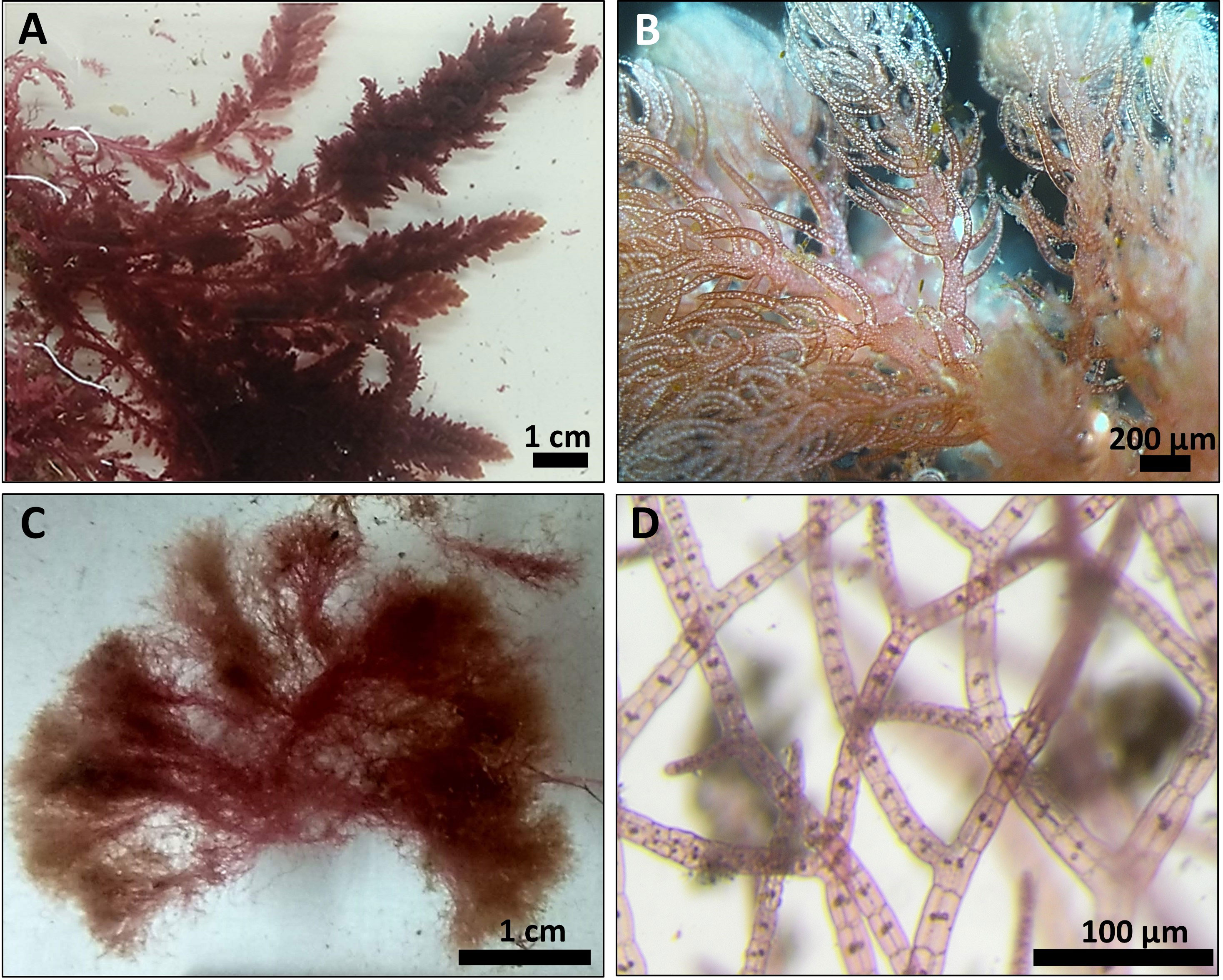
Figure 2 Typical A. taxiformis gametophyte collected from Tel Shikmona site (A, B) and tetrasporophyte collected from Rosh Hanikra site (C, D).
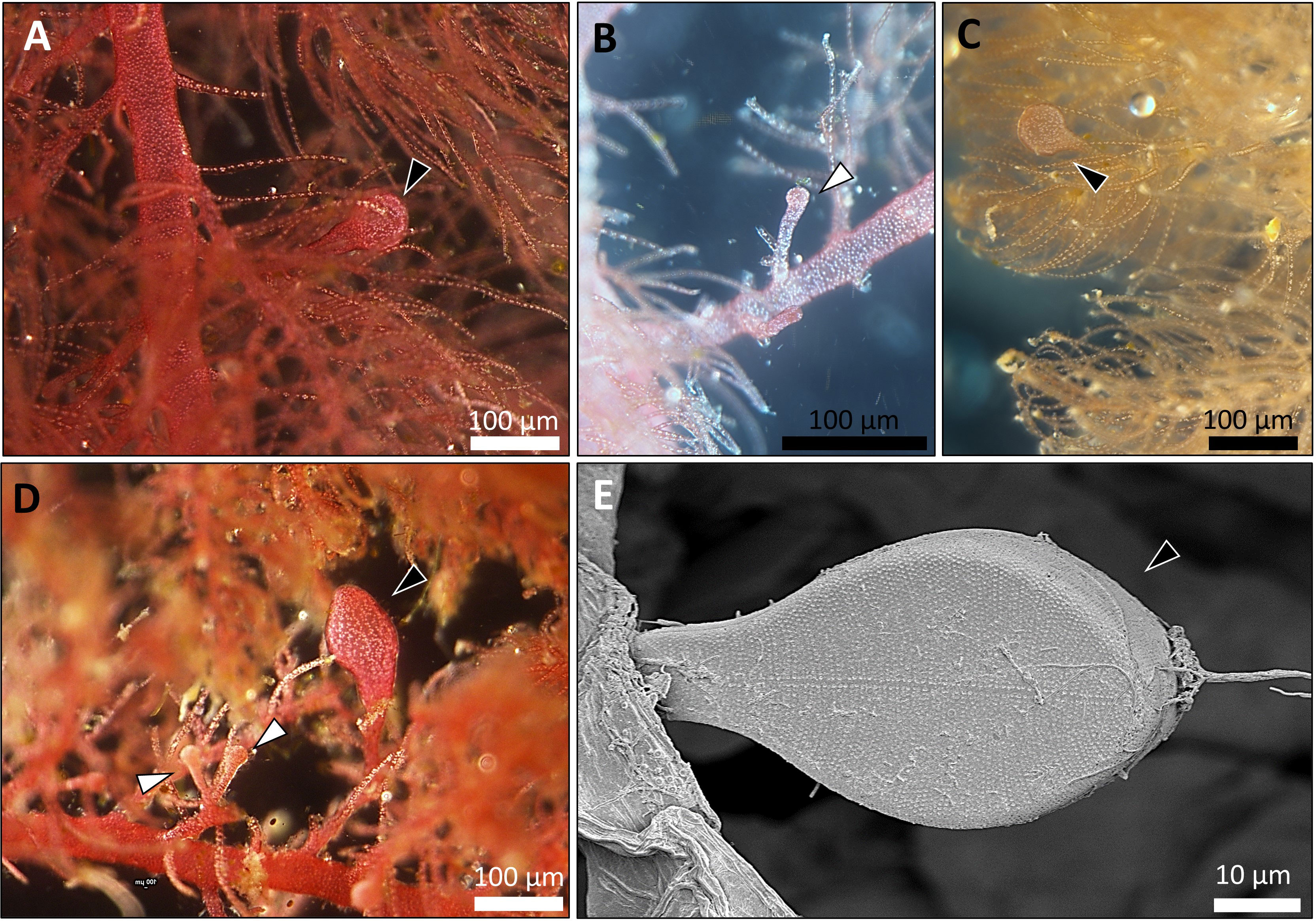
Figure 3 Fertile A. taxiformis from the IMS (photos of specimens correspond to samples collected during October 2021, from Tel Shikmona and Rosh Hanikra sites). (A–D) light microscope. (E) scanning electron microscope (SEM). Cystocarps (which indicate the development of the gonimoblast, the female reproductive structure) are shown with black arrow heads and spermatangium (male reproductive structure) by white arrow heads.
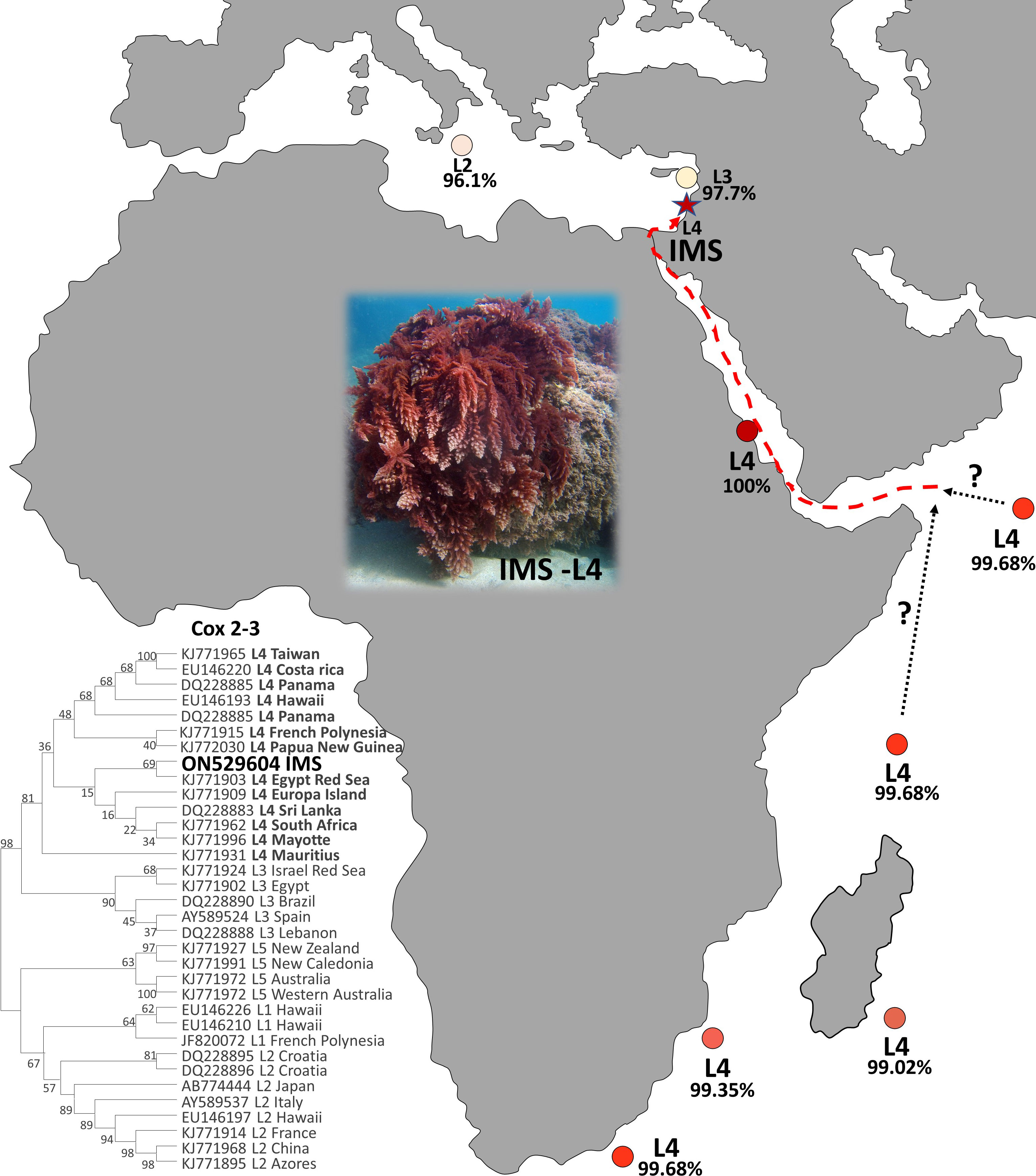
A total of 27 sequences were successfully generated for the cox2-3 spacer marker, 29 for the rbcL, and 23 for the LSU (Table S1). Phylogenetic trees constructed for the three different genes (Figure 4) revealed that all samples examined clustered within the lineage 4 (L4). All three markers, cox2-3 spacer, LSU and rbcL, show clustering with the IMS samples together with all L4 lineages samples deposited from GenBank. One exception is the LSU marker of the L4 sample (GenBank KJ772105) from the Egyptian Red Sea which clustered alone and may be a result of a single nucleotide polymorphism which is unique from all of the other samples and most likely may represent a sequencing error. Previous sequencing of the cox2-3 spacer from distant geographic sites such as Hawaii and Panama differ by 0.98% (3 out of 306 bp) from the IMS samples. Closer sequencing can be seen from L4 samples from French Polynesia and Papua New Guinea which have a discrepancy of 0.65% (2 out of 306 bp) from the IMS samples (Andreakis et al., 2007; Sherwood, 2008; Bolton et al., 2011; Dijoux et al., 2014; Andreakis et al., 2016; Kurihara et al., 2016). Samples from South Africa, Europa Island, and Sri Lanka diverged from the IMS samples in only one nucleotide (0.32%). The closest sequencing to the IMS samples is A. taxiformis L4 sampled from the Egyptian Red Sea with 100% similarity (Figure 5). In comparison to previously sequenced Mediterranean samples, the IMS cox2-3 marker differs by 2.3% and 3.9% from L3 and L2 Mediterranean populations, respectively (Andreakis et al., 2004; Andreakis et al., 2007; Dijoux et al., 2014) (Figure 5).
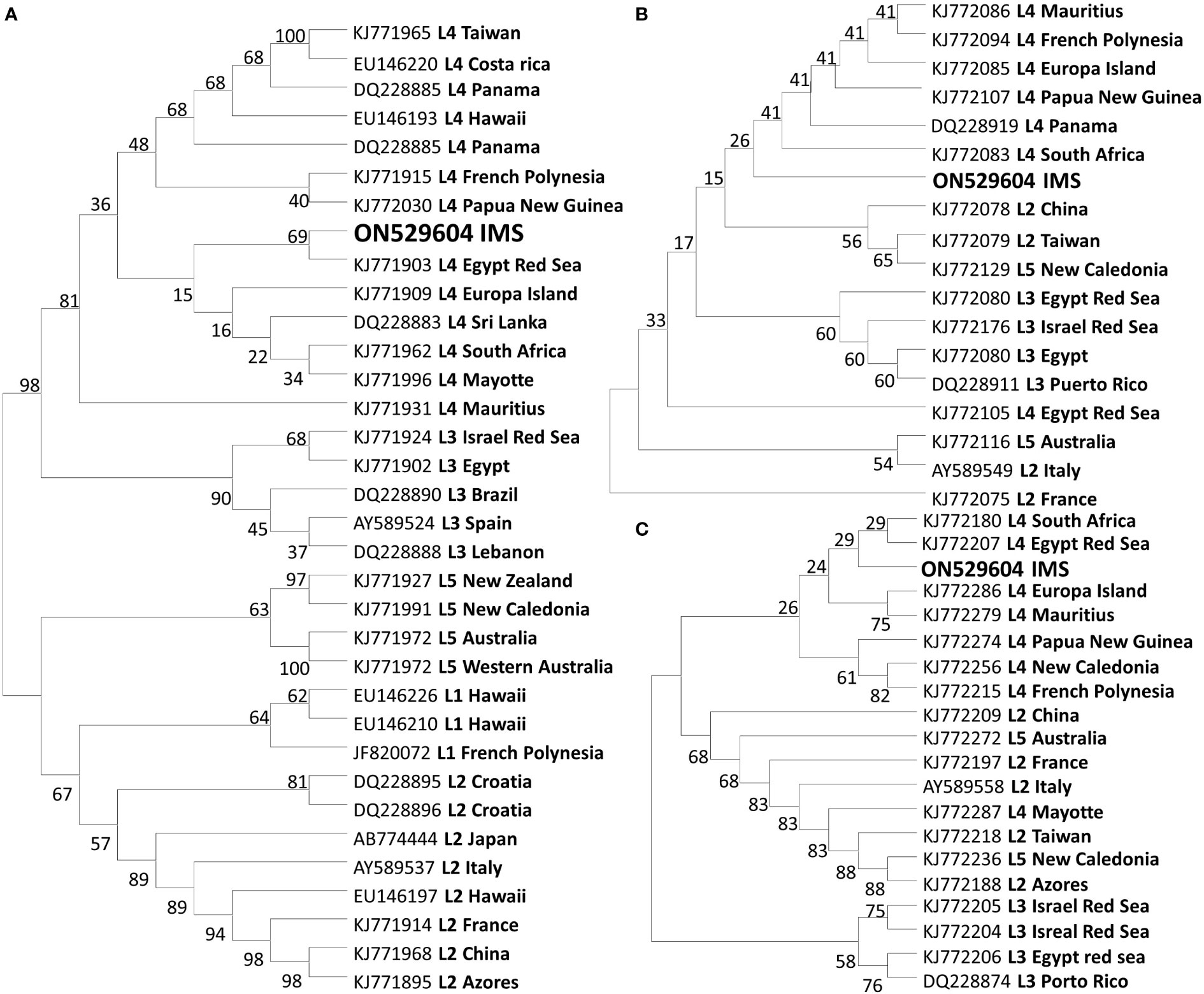
Figure 4 Evolutionary relationships trees of A taxiformis with the percentage of clustering according to the bootstrap test. Tree according to the cox2-3 spacer (A). Tree according to the LSU marker (B). Tree according to the rbcL marker (C).
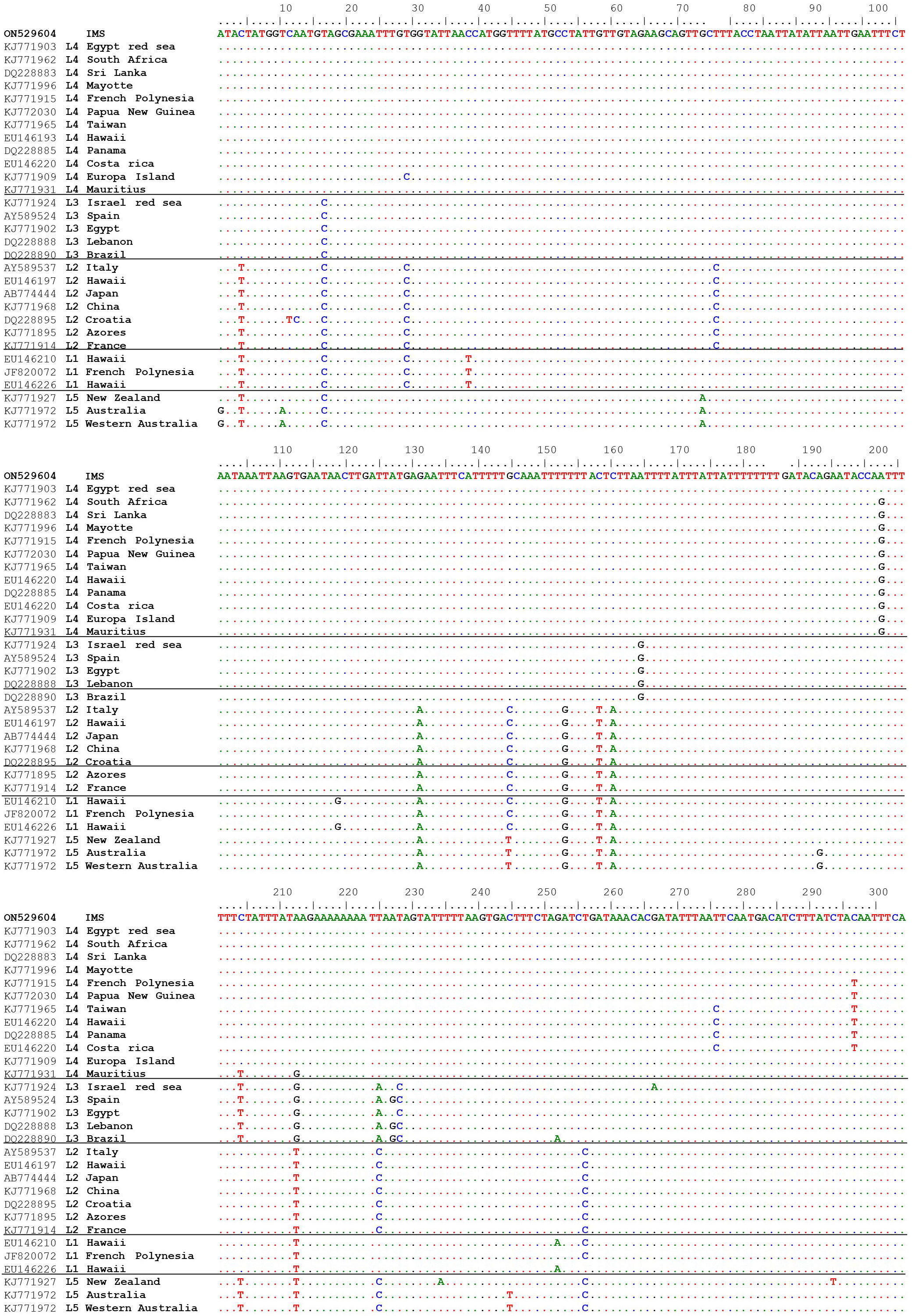
Figure 5 Multiple alignments of the cox2-3 spacer sequence isolated from IMS sample (GenBank accession number ON529604) compared to sequences from GenBank.
Discussion
The seaweed Asparagopsis taxiformis is an iconic example of macroalgal invasion from a global perspective, which has invaded almost all oceans and seas around the world (Zanolla et al., 2022). Until now, two cryptic lineages have been described for the Mediterranean Sea, L2 and L3 (Zanolla et al., 2022). In this study, we present solid evidence of an additional cryptic lineage (so-called L4) of A. taxiformis first reported for the Mediterranean Sea. We regard this lineage as relatively new when compared to the introduction of L2 and L3 genetic lines that were reported in the past. The wide geographic distribution of A. taxiformis L4 seems to be the result of long-distance, human-mediated dispersal events (Dijoux et al., 2014). The true native origin of L4 is unknown, however, the pathways of introduction into the Mediterranean Sea can be predicted. A soundly invasion route includes two distinct geographic origins, Sri Lanka or South Africa, both within the Indian Ocean. A. taxiformis populations may have established in the eastern Indian Ocean and reached the Red Sea, to eventually penetrate into the eastern Mediterranean Sea via Suez Canal. This possible route is supported by the similarity of the cox2-3 marker from the IMS sequences and between South Africa, Sri Lanka, and the Red Sea specimens (Figure 6). Whether A. taxiformis is a native species or has been introduced into the Mediterranean Sea is still unclear (Boudouresque and Verlaque, 2002; Zanolla et al., 2022). However, based on previous data collected on common Lessepsian migration route (Nunes et al., 2014) taken together with the proximity of the IMS to the Suez Canal, as well as the molecular data presented in this study, we hypothesize that A. taxiformis L4 is of a Lessepsian origin. From our observations and viewing the lack of ecological and molecular studies in the past, we suggest that A. taxiformis L4 introduction into the eastern Mediterranean Sea may have occurred only a few decades ago.
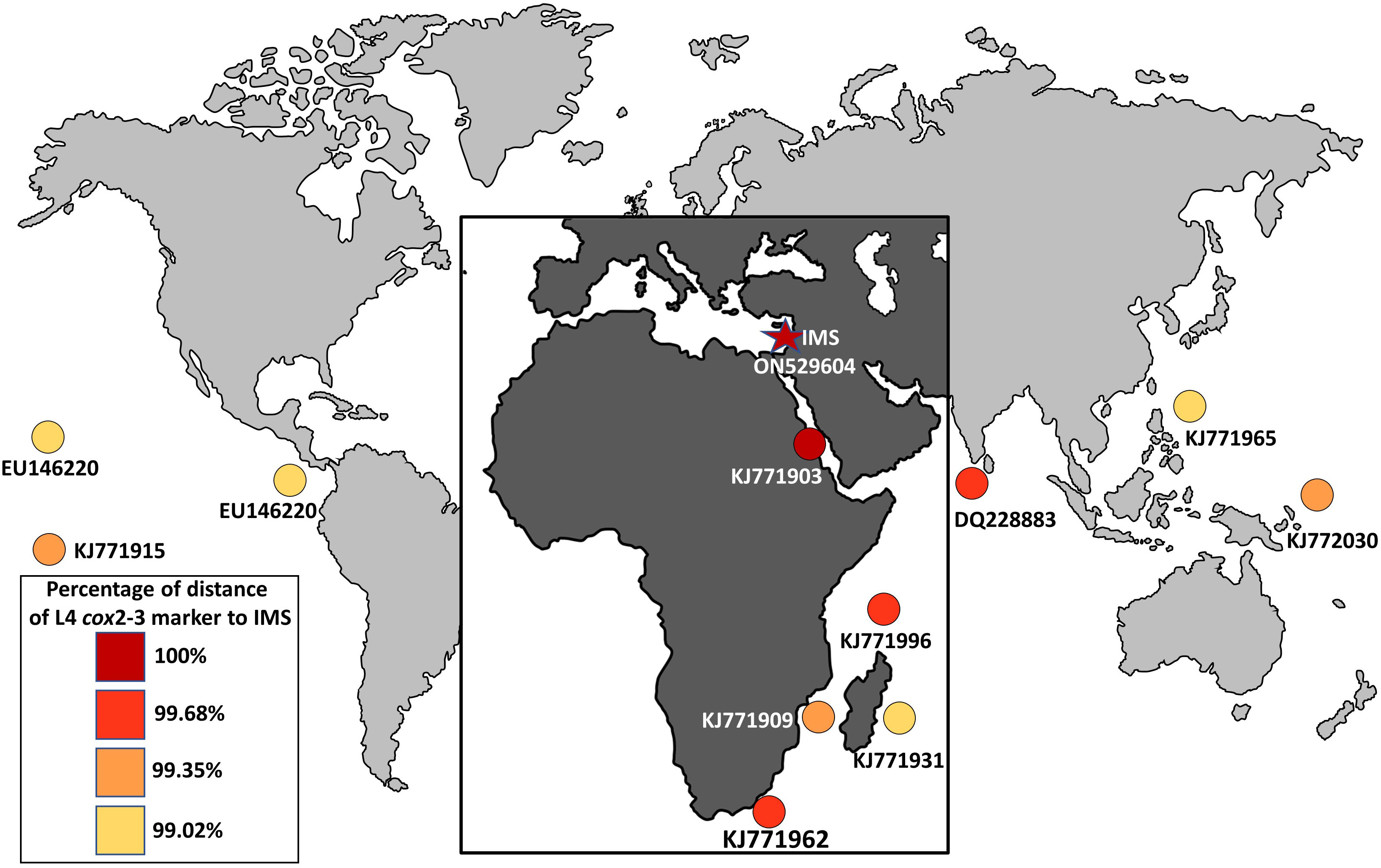
Figure 6 Global distribution of the lineage L4 (cox2-3 spacer marker) of A. taxiformis and their percentage of distance compared to the IMS samples in this study.
Morphological characters of A. taxiformis gametophyte have been used to distinguish between cryptic lineages (Zanolla et al., 2014). Furthermore, gametophytes from the same lineage may show different characters when comparing populations from different geographic regions. For instance, thallus height, basal diameter, and length of the branchlets were all greater in Mediterranean Sea invasive L2 as compared to Hawaiian L2 populations (Zanolla et al., 2014). The trend of increasing thallus morpho-parameters in the Mediterranean Sea populations compared to populations from the same lineage, but from distant geographic sites, was also seen in L4. Specifically, morphological traits of L4 from Rosh Hanikra site, namely thallus height, main axis basal diameter, and height where branching begins were higher compared to L4 from Hawaii. There is still uncertainty as to whether this tendency is due to the different biotic conditions in the Mediterranean Sea or whether the increased size of some specimens gives them ecological advantages, thus increasing their invasive rates and extent. Recently observed drifts strongly indicate that A. taxiformis is widespread in shallow habitats between Rosh Hanikra and Mikhmoret sites. In these sites, A. taxiformis is frequently attached to exposed rocky substrates both vertically and horizontally, usually between 0-5 m deep. Nonetheless, A. taxiformis is also quite abundant at depths of 20–30 m and significant drifts of fertile gametophytes are carried out to the shore following stormy sea days. We have no quantitative seasonal data for the above observations, yet our frequent field trips indicate that A. taxiformis exists all year round with productivity declining during wintertime. Growth capacity together with physiological and reproductive traits related to environmental factors for the IMS L4 A. taxiforims are still unknown. In a previous study which did not include genetic characterization, some populations of A. taxiformis were described as dioecious (Barone et al., 2013). As for the L4 lineage from Hawaii, Zanolla et al. did not report the presence of any reproductive structures (Zanolla et al., 2014). In this study, nearly every specimen showed reproductive structures carrying both cystocarps and spermatangia strongly indicating that IMS L4 is monoecious (Figure 3).
Detached gametophytes as seen in large amounts in the IMS could promote further dispersal of the L4 population in two possible ways. First, by vegetative reproduction via fragmentation of the thalli as suggested by Mairh (1977) and second, although the released gametes and tetraspores are not thought to travel very far (Santelices, 1990), detached gametophytes that drift with the currents can carry and release reproductive structures for long distances. The presence of the gametophyte in a site does not necessarily confirm the presence of the tetrasporophytes stage, and vice versa (Bolton et al., 2011; Orlando-Bonaca et al., 2017). The observation of both reproductive stages in the same site indicates an active reproductive population. Furthermore, the occurrence of tetrasporophytes, which are known as a good dispersal unit, makes this population a potential donor source for future invasive populations
All A. taxiformis specimens analyzed from the Mediterranean Sea so far were found to be either L2 or L3 (Zanolla et al., 2022). Studies involving the population dynamics of the Mediterranean L2 suggest the successful expansion of this lineage can be explained by the temperate and tropical preferences of this lineage (Zanolla et al., 2018). Furthermore, rising sea temperatures in some regions due to climate change will also increase the opportunity for this lineage to further spread in those areas (Zanolla et al., 2018). Zanolla et al. (2018) also highlight the co-existence of L2 and A. armata in some areas, suggesting as the former existence of A. armata promotes the L2 expansion in those sites. This conjecture relies on the assumption that communities under stress from an invasive species are more susceptible to additional invaders (Simberloff and Von Holle, 1999). Considering that A. taxiformis is already well established in almost all of the Mediterranean Sea, the ability of L4 the flourish in tropical-temperate regions, and the suggested Lessepsian migration route, there is a significant likelihood that L4 populations will spread to other Mediterranean basins. Moreover, considering the classic Lessepsian migration route of NISS we hypothesize Tsiamis and Panayotidis (2007) first description of A. taxiformis in Greece (which not include molecular work), might have corresponded to the L4 described in this study. The increasing abundance of A. taxiformis L4 in the eastern Mediterranean Sea underlines the need for a comprehensive monitoring strategy for its distribution and lineage description. Lastly, analyses of macroalgae herbaria samples may be crucial to assess their impact on local biodiversity and track their possible route over time.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
All authors design the research and conduct the field collection. ON did the molecular work and data analysis. All authors provided critical feedback and helped shape the research, analysis, and manuscript. All authors read and approved the final manuscript.
Funding
This project was funded by the Ph.D. excellence scholarshipof the University of Haifa given to ON, and by Ministry ofHealth, Israel grant no. 3-16052 given to AI.
Acknowledgments
We are thankful to Guy Paz for preparing the artwork. In addition, we want to thank Dr. Maya Lalzar from the University of Haifa Bioinformatics Service Unit for her assistance and Dr. Dikla Aharonovich for her help with electron microscopy.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.873817/full#supplementary-material
References
Agency E (2007). Halting the loss of biodiversity by 2010: Proposal for a first set of indicators to monitor progress in Europe (Technical Report 11/2007: European Environment Agency - EEA (2007).
Andreakis N., Costello P., Zanolla M., Saunders G. W., Mata L. (2016). Endemic or introduced? phylogeography of Asparagopsis (Florideophyceae) in Australia reveals multiple introductions and a new mitochondrial lineage. J. Phycol. 52, 141–147. doi: 10.1111/jpy.12373
Andreakis N., Procaccini G., Kooistra W. H. C. F. (2004). Asparagopsis taxiformis and Asparagopsis armata (Bonnemaisoniales, rhodophyta): Genetic and morphological identification of Mediterranean populations. eur. J. Phycol . 39, 273–283. doi: 10.1080/0967026042000236436
Andreakis N., Procaccini G., Maggs C., Kooistra W. H. C. F. (2007). Phylogeography of the invasive seaweed Asparagopsis (Bonnemaisoniales, rhodophyta) reveals cryptic diversity. Mol. Ecol. 16, 2285–2299. doi: 10.1111/j.1365-294X.2007.03306.x
Andreakis N., Schaffelke B. (2012). ““Invasive marine seaweeds: Pest or prize?,”,” in Seaweed biology, 471–493 . Springer, New York. doi: 10.1007/978-3-642-28451-9
Barone R., Mannino A. M., Marino M. (2013). Asparagopsis taxiformis (Bonnemaisoniales, rhodophyta): first record of gametophytes on the Italian coast. Bocconea 16, 1021–1025.
Bickford D., Lohman D. J., Sodhi N. S., Ng P. K. L., Meier R., Winker K., et al. (2007). Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 22, 148–155. doi: 10.1016/j.tree.2006.11.004
Bolton J. J., Andreakis N., Anderson R. J. (2011). Molecular evidence for three separate cryptic introductions of the red seaweed Asparagopsis ( bonnemaisoniales , rhodophyta ) in south Africa. Afr. J. Mar. Sci. 33, 263–271. doi: 10.2989/1814232X.2011.600339
Boni D. R., Hawkes M. W. (1987). Systematics and life histories of new Zealand bonnemaisoniaceae (Bonnemaisoniales, rhodophyta): I. the genus Asparagopsis. New Zeal. J. Bot. 25, 577–590. doi: 10.1080/0028825X.1987.10410088
Boudouresque C. F., Verlaque M. (2002). Biological pollution in the Mediterranean Sea: invasive versus introduced macrophytes. Biol. Invasions 14, 32–38. doi: 10.1016/S0025-326X(01)00150-3
Boudouresque C. F., Verlaque M. (2005). Nature conservation , marine protected areas , sustainable development and the flow of invasive species to the Mediterranean Sea. Sci. Rep. du Parc Natl. PortCros 54, 29–54.
Chihara M. (1961). Life cycle of bonnemaisoniaceous algae in Japan. Sci. Rep. Tokyo Kyoiku Daiq. Sect. B 10, 121–154.
Dijoux L., Viard F., Payri C. (2014). The more we search , the more we Find : Discovery of a new lineage and a new species complex in the genus Asparagopsis. PLos One 9, e103826. doi: 10.1371/journal.pone.0103826
Einav R., Israel A. (2008). Checklist of seaweeds from the Israeli Mediterranean: Taxonomical and ecological approaches. isr. J. Plant Sci. 56, 127–191. doi: 10.1560/IJPS.57.1
Einav R., Israel A. (2009). Stypopodium schimperi. another brown algal bloom. Teva Hadvarim Hebr 11:165.
FD P. (1978). Lessepsian migration: the influx of red Sea biota into the Mediterranean by way of the Suez canal (New York: Springer Berlin Heidelberg).
Grosholz E. (2002). Ecological and evolutionary consequences of coastal invasions. Trends Ecol. Evol. 17, 22–27. doi: 10.1016/S0169-5347(01)02358-8
Guiry M. D., Dawes C. J. (1992). Daylength, temperature and nutrient control of tetrasporogenesis in Asparagopsis armata (Rhodophyta). J. Exp. Mar. Bio. Ecol. 158, 197–217. doi: 10.1016/0022-0981(92)90227-2
Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. 41, 95–98.
Harvey W. H. (1849). Some account of the marine botany of the colony of Western Australia. Trans. R. Irish Acad. 22, 525–566.
Hoffman R., Israel A., Lipkin Y., Dubinsky Z., Iluz D. (2008). First record of two seaweeds from the Israeli Mediterranean: Galaxaura rugosa (J. Ellis and solander) j. v. lamouroux (Rhodophyta) and Codium adhaerens c. agardh (Chlorophyta). Isr. J. Plant Sci. 2, 123–126.
Israel A., Einav R., Silva P. C., Paz G., Chacana M. E., Douek J. (2010). First report of the seaweed Codium parvulum (Chlorophyta) in Mediterranean waters: Recent blooms on the northern shores of Israel. Phycologia 49, 107–112. doi: 10.2216/PH09-28.1
Israel A., Golberg A., Neori A. (2019). The seaweed resources of Israel in the Eastern Mediterranean Sea. Bot. Mar 63, 85–95. doi: 10.1515/bot-2019-0048
Katsanevakis S., Coll M., Piroddi C., Steenbeek J., Lasram F. B. R., Zenetos A., et al. (2014). Invading the Mediterranean Sea: Biodiversity patterns shaped by human activities. Front. Mar. Sci. 1. doi: 10.3389/fmars.2014.00032
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kurihara A., Horiguchi H., Hanyuda T., Kawai H. (2016). Phylogeography of Asparagopsis taxiformis revisited: Combined mtDNA data provide novel insights into population structure in Japan. Phycol. Res. 64, 95–101. doi: 10.1111/pre.12126
Lenaers G., Michot L. M. B., Herzog M. (1989). Dinoflagellates in evolution. a molecular phylogenetic analysis of Large subunit ribosomal RNA 29, 40–51.
Lipkin Y. (1962). “Ecological observations on the mikhmoret coast,” in Preliminary survey of the estival aspect of the marine vegetation. (in Hebrew, English abstr.) (Jerusalem:Department of Botany: Hebrew University).
Maggs C. A., Douglas S. E., Fenety J., Bird C. J. (1992). A molecular and morphological analysis of the Gymnogongrus devoniensis (Rhodophyta) complex in the north Atlantic. J. Phyco 28, 214–232. doi: 10.1111/j.0022-3646.1992.00214.x
Mairh O. P. (1977). Studies on Asparagopsis taxiformis delile Collins and Harvey from Indian waters. J. Mar. Biol. Assoc. India 96, 97–106.
Ní Chualáin F., Maggs C. A., Saunders G. W., Guiry M. D. (2004). The invasive genus Asparagopsis (Bonnemaisoniaceae, rhodophyta): Molecular systematics, morphology, and ecophysiology of falkenbergia isolates. J. Phycol. 40, 1112–1126. doi: 10.1111/j.1529-8817.2004.03135.x
No Nei M., Kumar S. (2000). Molecular evolution and phylogenetics (New York: Oxford University Press).
Nunes A. L., Katsanevakis S., Zenetos A., Cardoso A. C. (2014). Gateways to alien invasions in the European seas. Aquat. Invasions 9, 133–144. doi: 10.3391/ai.2014.9.2.02
Orlando-Bonaca M., Mavrič B., Trkov D., Lipej L. (2017). Unusual bloom of tetrasporophytes of the non-indigenous red alga Asparagopsis armata in the northern Adriatic sea. Acta Adriat. 58, 53–62. doi: 10.32582/aa.58.1.4
Orsini L., Sarno D., Procaccini G., Poletti R., Dahlmann J., Montresor M. (2002). Toxic Pseudo-nitzschia multistriata (Bacillariophyceae) from the gulf of Naples: Morphology, toxin analysis and phylogenetic relationships with other pseudo-nitzschia species. Eur. J. Phycol. 37, 247–257. doi: 10.1017/S0967026202003608
Price J., John D. M., Lawson G. M. (1986). Seaweeds of the western coast of tropical Africa and adjacent islands: a critical assessment IV. rhodophyta (Florideae). Bull. Br. Mus 1, 1–122.
Rilov G., Crooks J. A. (2009). Biological invasions in marine ecosystems (Berlin, Germany: Springer).
Rojas J. J., Lemus A., Ganesan E. K. (1982). El Ciclo vital “in vitro” del alga marina roja Asparagopsis taxiformis (Delile) Collins & hervey (Bonnemaisoniales, rhodophyta). del Mar. Caribe. Boll. Inst. Ocean. 21, 101–112.
Romero J., Alcoverro T., Roca G., Pérez M. (2016). Bioindicators, monitoring, and management using Mediterranean seagrasses: What have we learned from the implementation of the EU water framework directive? (Part II) Vol. vol 43. Eds. Munné A., Ginebreda A., Prat N. (Cham: Springer).
Santelices B. (1990). Patterns of reproduction, dispersal and recruitment in seaweeds. Oceanogr. Mar. 28, 177–276.
Sherwood A. R. (2008). Phylogeography of Asparagopsis taxiformis (Bonnemaisoniales, rhodophyta) in the Hawaiian islands: Two mtDNA markers support three separate introductions. Phycologia 47, 79–88. doi: 10.2216/07-39.1
Silva P. C., Basson P. W., Moe R. L. (1996). Catalogue of the benthic marine algae of the Indian ocean. Berkeley and Los Angeles University of California publications in botany. (Berkeley: University of California Press)
Simberloff D., Von Holle B. (1999). Positive interactions of nonindigenous species: Invasional meltdown? Biol. Invasions 1, 21–32. doi: 10.1023/A:1010086329619
Streftaris N., Zenetos A. (2006). Alien marine species in the Mediterranean - the 100 ‘Worst invasives’ and their impact. Mediterr. Mar. Sci. 7, 87–118. doi: 10.12681/mms.180
Tsiamis K., Panayotidis P. (2007). First record of the red alga Asparagopsis taxiformis (Delile) trevisan de saint-léon in Greece. Aquat. Invasions 2, 435–438. doi: 10.3391/ai.2007.2.4.14
Verlaque M., Boudouresque C. (1991). Stypopodium schimperi (Buchinger ex kützing) verlaque et boudouresque comb. nov. (Dictyotales, fucophyceaea), algue de mer rouge récemment apparue en méditerranée. Cryptogam. Algol. 12, 195–211.
Verlaque M., Ruitton S., Mineur F., Boudouresque C.-F. (2015). CIESM atlas of exotic species in the Mediterranean Vol. 4 (Macrophytes. Monaco: CIESM Publishers).
Vieira C., Aharonov A., Paz G., Engelen A. H., Tsiamis K., Einav R., et al. (2019). Diversity and origin of the genus Lobophora in the Mediterranean Sea including the description of two new species. Phycologia 58, 163–168. doi: 10.1080/00318884.2018.1534923
Zanolla M., Altamirano M., Carmona R., de la Rosa J., Souza-Egipsy V., Sherwood A., et al. (2018). Assessing global range expansion in a cryptic species complex: insights from the red seaweed genus Asparagopsis (Florideophyceae). J. Phycol. 54, 12–24. doi: 10.1111/jpy.12598
Zanolla M., Carmona R., de la Rosa J., Salvador N., Sherwood A. R., Andreakis N., et al. (2014). Morphological differentiation of cryptic lineages within the invasive genus Asparagopsis (Bonnemaisoniales, rhodophyta). Phycologia 53, 233–242. doi: 10.2216/13-247.1
Zanolla M., Carmona R., Mata L., de la Rosa J., Sherwood A., Barranco C. N., et al. (2022). Concise review of the genus Asparagopsis montagne 1840. J. Appl. Phycol. 34, 1–17. doi: 10.1007/s10811-021-02665-z
Zenetos A. (2010). Trend in aliens species in the mediterranean. an answer to galil 2009 “Taking stock: Inventory of alien species in the Mediterranean sea.” Biol. Invasions 12, 3379–3381. doi: 10.1007/s10530-009-9679-x
Zenetos A., Gofas S., Morri C., Rosso A., Violanti D., García Raso J. E., et al. (2012). Alien species in the Mediterranean Sea by 2012. a contribution to the application of European union’s marine strategy framework directive (MSFD). part 2. introduction trends and pathways. Mediterr. Mar. Sci. 13, 328–352. doi: 10.12681/mms.327
Keywords: Asparagopsis taxiformis, cox2-3 spacer, Lessepsian migration, Mediterranean Sea, seaweed
Citation: Nahor O, Luzzatto-Knaan T and Israel Á (2022) A new genetic lineage of Asparagopsis taxiformis (Rhodophyta) in the Mediterranean Sea: As the DNA barcoding indicates a recent Lessepsian introduction. Front. Mar. Sci. 9:873817. doi: 10.3389/fmars.2022.873817
Received: 11 February 2022; Accepted: 28 June 2022;
Published: 28 July 2022.
Edited by:
Stelios Katsanevakis, University of the Aegean, GreeceReviewed by:
Marianela Zanolla, University of Malaga, SpainSimona Armeli Minicante, Institute of Marine Science (CNR), Italy
Copyright © 2022 Nahor, Luzzatto-Knaan and Israel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tal Luzzatto-Knaan, tluzzatto@univ.haifa.ac.il; Alvaro Israel, alvaro@ocean.org.il
 Omri Nahor
Omri Nahor Tal Luzzatto-Knaan
Tal Luzzatto-Knaan Álvaro Israel
Álvaro Israel