
Read the latest Issue
EMBL researchers combine multiple datasets to develop expandable atlas of an entire animal
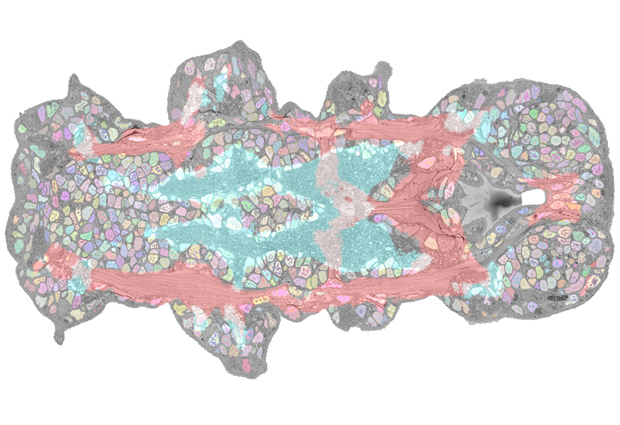
The segmented marine worm Platynereis dumerilii is an ideal organism for the study of the nervous system. At EMBL Heidelberg, researchers have developed a resource that enables completely new insights into the biology of this organism. PlatyBrowser is an interactive atlas of the entire animal that allows scientists to travel virtually to any cell found in a young Platynereis worm, inspect the cell’s ultrastructure in its anatomical context, and display the genes that are expressed in that cell. The resource and all the data have been made available both online and in a preprint. The researchers invite feedback from users, who can also add their own datasets to the atlas.
The novel Platynereis atlas – a collaborative project involving researchers from EMBL’s Arendt and Kreshuk groups, Schwab team and Electron Microscopy Core Facility, and Centre for Bioimage Analysis – is based on volume electron microscopy (EM) data acquired from a whole six-day-old worm. “We acquired high-resolution volume EM data of an entire animal, which provides ultrastructural detail,” explains Detlev Arendt, EMBL group leader and one of the coordinators of the project. “We’re also bringing together spatial expression profiling and volume EM for the first time at this scale.”
The study was made possible by recent advances in EM technology and data analysis. In collaboration with Christel Genoud and Rainer Friedrich at the Friedrich Miescher Institute for Biomedical Research (FMI) in Basel, Switzerland, who have developed a new software tool called SBEMimage, the researchers managed to acquire the several terabytes of data required to image a complete specimen at nanometre resolution. Novel algorithms were used to automatically segment all 12 000 cells and their nuclei across all tissues in Platynereis. “Developments in the field of connectomics, which studies neuronal structures by EM, pioneered these methods, but we had to develop new approaches for segmentation of very diverse tissues across the whole organism, which is really challenging,” explains EMBL group leader Anna Kreshuk. “So far, nobody had done this for non-neuronal tissues or multiple tissue types at once.”
The nuclear segmentation pattern could then be used to align the EM data with a previously published and expanded gene expression dataset for hundreds of genes. This enables users to zoom in on single cells to simultaneously investigate their structure, composition, and the genes expressed within them. “We now have a full overlay of gene expression data onto an anatomical atlas at nanometric resolution,” says EMBL team leader Yannick Schwab. “This is the first time such a detailed match has been obtained for an entire organism.”
To make the combined gene expression and EM data easily explorable, the team has developed a new Fiji plugin that allows browsing of large multimodal datasets. “We collaborated with EMBL’s IT department to use the latest developments in cloud data storage to enable remote public access to terabyte-sized image datasets,” explains Christian Tischer, who runs EMBL’s Centre for Bioimage Analysis. The PlatyBrowser plugin is available for anyone to install as open source software.
The current research project is a team effort across EMBL units and research groups, as well as with external collaborators. It unites expertise in Platynereis biology, EM technology, automated image processing, and machine learning from the Arendt and Kreshuk groups, the Schwab team, EMBL’s Centre for Bioimage Analysis, and the collaborators at FMI Basel. Bringing together this expertise has not only allowed the researchers to overcome the technical challenges, but has already revealed novel insights into Platynereis biology. “We can now cluster segmented cells that show similar gene expression, and we find that these clusters follow morphological boundaries in the animal’s body. There is a high coherence between gene expression at the tissue level and the anatomical units in the body. This type of insight has not been possible before,” says Arendt.
The biggest strength of the new resource, though, is its multi-modality. “This means that several types of data can be combined,” Arendt explains. “At the moment, we have gene expression and the ultrastructure, but our resource allows other modalities, such as metabolomics or proteomics datasets, or X-ray tomography data to be easily added.” This way, PlatyBrowser enables researchers to perform multidisciplinary analyses and add their own research data. The EMBL researchers invite the whole Platynereis community to use the resource and provide feedback. “Our resource is expandable; it’s not a fixed resource,” Schwab concludes. “It will keep growing with more knowledge being added to it in the future.”
Looking for past print editions of EMBLetc.? Browse our archive, going back 20 years.
EMBLetc. archive