Time to reexamine some assumptions (again)! And also, talk about Hox genes, because do I even need a reason?
Hox genes often come up when we look for explanations for various innovations in animal body plans – the digits of land vertebrates, the limbless abdomens of insects, the various feeding and walking and swimming appendages of crustaceans, the strongly differentiated vertebral columns of mammals, and so on.
Speaking of differentiated vertebral columns, here’s one group I’d always thought of as having pretty much the exact opposite of them: snakes. Vertebral columns are patterned, among other things, by Hox genes. Boundaries between different types of vertebrae such as cervical (neck) and thoracic (the ones bearing the ribcage) correspond to boundaries of Hox gene expression in the embryo – e.g. the thoracic region in mammals begins where HoxC6 starts being expressed.
In mammals like us, and also in archosaurs (dinosaurs/birds, crocodiles and extinct relatives thereof), these boundaries can be really obvious and sharply defined – here’s Wikipedia’s crocodile skeleton for an example:

In contrast, the spine of a snake (example from Wikipedia below) just looks like a very long ribcage with a wee tail:

Snakes, of course, are rather weird vertebrates, and weird things make us sciencey types dig for an explanation.
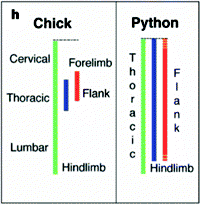
Since Hox genes appear to be responsible for the regionalisation of vertebral columns in mammals and archosaurs, it stands to reason that they’d also have something to do with the comparative lack of regionalisation (and the disappearance of limbs) seen in snakes and similar creatures. In a now classic paper, Cohn and Tickle (1999) observed that unlike in chicks, the Hox genes that normally define the neck and thoracic regions are kind of mashed together in embryonic pythons. Below is a simple schematic from the paper showing where three Hox genes are expressed along the body axis in these two animals. (Green is HoxB5, blue is C8, red is C6.)
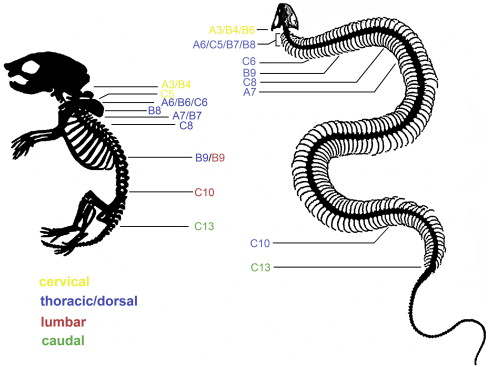
As more studies examined snake embryos, others came up with different ideas about the patterning of serpentine spines. Woltering et al. (2009) had a more in-depth look at Hox gene expression in both snakes and caecilians (limbless amphibians) and saw that there are in fact regions ruled by different Hoxes in these animals, if a little fuzzier than you’d expect in a mammal or bird – but they don’t appear to translate to different anatomical regions. Here’s their summary of their findings, showing the anteriormost limit of the activity of various Hox genes in a corn snake compared to a mouse:
Such differences aside, both of the above studies operated on the assumption that the vertebral column of snakes is “deregionalised” – i.e. that it evolved by losing well-defined anatomical regions present in its ancestors. But is that actually correct? Did snakes evolve from more regionalised ancestors, and did they then lose this regionalisation?
Head and Polly (2015) argue that the assumption of deregionalisation is a bit stinky. First, that super-long ribcage of snakes does in fact divide into several regions, and these regions respect the usual boundaries of Hox expression. Second, ordinary lizard-shaped lizards (from which snakes descended back in the days of the dinosaurs) are no more regionalised than snakes.
The study is mostly a statistical analysis of the shapes of vertebrae. Using an approach called geometric morphometrics, it turned these shapes from dozens of squamate (snake and lizard) species into sets of coordinates, which could then be compared to see how much they vary along the spine and whether the variation is smooth and continuous or clustered into different regions. The authors evaluated hypotheses regarding the number of distinct regions to see which one(s) best explained the observed variation. They also compared the squamates to alligators (representing archosaurs).
The results were partly what you’d expect. First, alligators showed much more overall variation in vertebral shape than squamates. Note that that’s all squamates – leggy lizards are nearly (though not quite) as uniform as their snake-like relatives. However, in all squamates, the best-fitting model of regionalisation was still one with either three or four distinct regions in front of the hips/cloaca, and in the majority, it was four, the same number as the alligator had.
Moreover, there appeared to be no strong support for an evolutionary pattern to the number of regions – specifically, none of the scenarios in which the origin of snake-like body plans involved the loss of one or more regions were particularly favoured by the data. There was also no systematic variation in the relative lengths of various regions; the idea that snakes in general have ridiculously long thoraxes is not supported by this analysis.
In summary, snakes might show a little less variation in vertebral shape than their closest relatives, but they certainly didn’t descend from alligator-style sharply regionalised ancestors, and they do still have regionalised spines.
Hox gene expression is not known for most of the creatures for which vertebral shapes were analysed, but such data do exist for mammals (mice, here), alligators, and corn snakes. What is known about different domains of Hox gene activation in these three animals turns out to match the anatomical boundaries defined by the models pretty well. In the mouse and alligator, Hox expression boundaries are sharp, and the borders of regions fall within one vertebra of them.
In the snake, the genetic and morphological boundaries are both gradual, but the boundaries estimated by the best model are always within the fuzzy boundary region of an appropriate Hox gene expression domain. Overall, the relationship between Hox genes and regions of the spine is pretty consistent in all three species.
To finish off, the authors make the important point that once you start turning to the fossil record and examining extinct relatives of mammals, or archosaurs, or squamates, or beasties close to the common ancestor of all three groups (collectively known as amniotes), you tend to find something less obviously regionalised than living mammals or archosaurs – check out this little figure from Head and Polly (2015) to see what they’re talking about:
(Moving across the tree, Seymouria is an early relative of amniotes but not quite an amniote; Captorhinus is similarly related to archosaurs and squamates, Uromastyx is the spiny-tailed lizard, Lichanura is a boa, Thrinaxodon is a close relative of mammals from the Triassic, and Mus, of course, is everyone’s favourite rodent. Note how alligators and mice really stand out with their ribless lower backs and suchlike.)
Although they don’t show stats for extinct creatures, Head and Polly argue that mammals and archosaurs, not snakes, are the weird ones when it comes to vertebral regionalisation. For most of amniote evolution, the norm was the more subtle version seen in living squamates. It was only during the origin of mammals and archosaurs that boundaries were sharpened and differences between regions magnified. Nice bit of convergent/parallel evolution there!
***
References:
Cohn MJ & Tickle C (1999) Developmental basis of limblessness and axial patterning in snakes. Nature 399:474-479
Head JJ & Polly PD (2015) Evolution of the snake body form reveals homoplasy in amniote Hox gene function. Nature 520:86-89
Woltering JM et al. (2009) Axial patterning in snakes and caecilians: evidence for an alternative interpretation of the Hox code. Developmental Biology 332:82-89